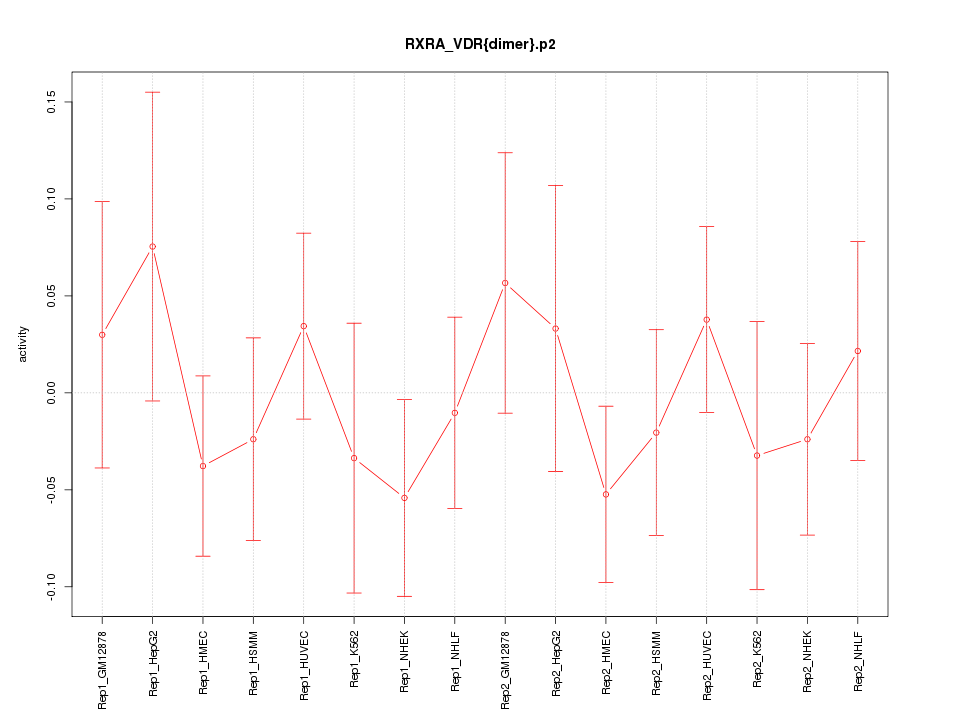
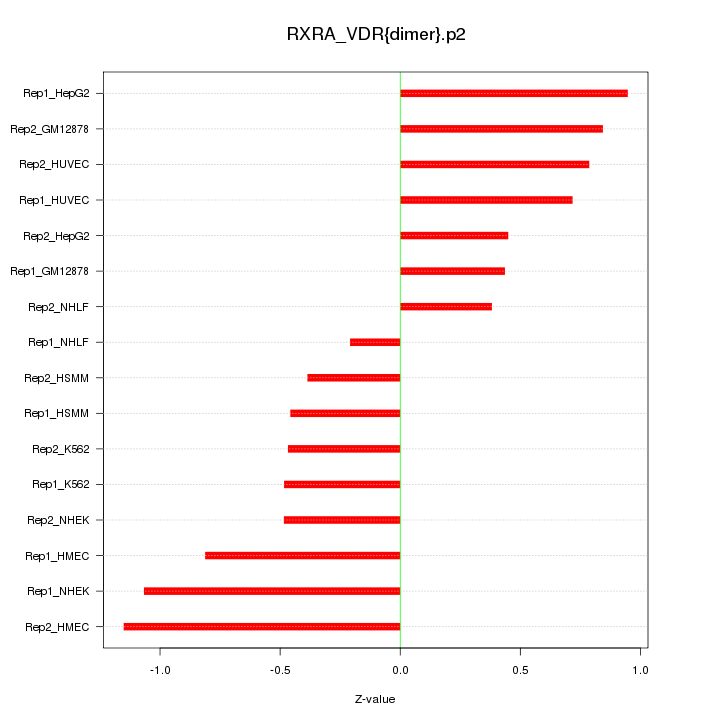
|
chr14_+_94117483
|
1.089
|
NM_000624
|
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5
|
|
chr7_+_94374862
|
1.044
|
NM_001166160
NM_017650
|
PPP1R9A
|
protein phosphatase 1, regulatory (inhibitor) subunit 9A
|
|
chr10_+_102722275
|
1.009
|
NM_017893
|
SEMA4G
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G
|
|
chr1_+_99884018
|
0.812
|
NM_017734
|
PALMD
|
palmdelphin
|
|
chr1_+_199259664
|
0.728
|
|
|
|
|
chr17_-_61655915
|
0.708
|
NM_000042
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I)
|
|
chr19_-_7244876
|
0.629
|
NM_000208
NM_001079817
|
INSR
|
insulin receptor
|
|
chr6_-_132314004
|
0.624
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr11_+_64620276
|
0.576
|
|
C11orf2
|
chromosome 11 open reading frame 2
|
|
chr11_+_64620287
|
0.571
|
|
C11orf2
|
chromosome 11 open reading frame 2
|
|
chr14_+_94117511
|
0.548
|
|
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5
|
|
chr11_+_64620183
|
0.541
|
NM_013265
|
C11orf2
|
chromosome 11 open reading frame 2
|
|
chr16_+_18986459
|
0.538
|
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr17_-_31831093
|
0.511
|
NM_001123392
NM_032258
|
TBC1D3C
TBC1D3G
TBC1D3
TBC1D3H
TBC1D3F
|
TBC1 domain family, member 3C
TBC1 domain family, member 3G
TBC1 domain family, member 3
TBC1 domain family, member 3H
TBC1 domain family, member 3F
|
|
chr17_+_18320822
|
0.496
|
NM_001040078
|
LGALS9B
LGALS9C
|
lectin, galactoside-binding, soluble, 9B
lectin, galactoside-binding, soluble, 9C
|
|
chr17_-_33602374
|
0.493
|
NM_032258
|
TBC1D3C
TBC1D3G
TBC1D3
TBC1D3F
|
TBC1 domain family, member 3C
TBC1 domain family, member 3G
TBC1 domain family, member 3
TBC1 domain family, member 3F
|
|
chr17_-_33552166
|
0.486
|
NM_001123391
NM_032258
|
TBC1D3C
TBC1D3G
TBC1D3
TBC1D3F
|
TBC1 domain family, member 3C
TBC1 domain family, member 3G
TBC1 domain family, member 3
TBC1 domain family, member 3F
|
|
chr17_-_31528027
|
0.429
|
NM_001001417
|
TBC1D3B
TBC1D3C
TBC1D3G
TBC1D3F
|
TBC1 domain family, member 3B
TBC1 domain family, member 3C
TBC1 domain family, member 3G
TBC1 domain family, member 3F
|
|
chr21_-_46399900
|
0.418
|
NM_006657
NM_206965
|
FTCD
|
formiminotransferase cyclodeaminase
|
|
chr17_-_31616071
|
0.418
|
NM_001001418
NM_032258
|
TBC1D3B
TBC1D3C
TBC1D3G
TBC1D3F
|
TBC1 domain family, member 3B
TBC1 domain family, member 3C
TBC1 domain family, member 3G
TBC1 domain family, member 3F
|
|
chr17_-_7023378
|
0.412
|
NM_001197216
NM_001671
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chrX_+_153423652
|
0.408
|
NM_001099856
|
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma
|
|
chr2_+_20510312
|
0.369
|
NM_004040
|
RHOB
|
ras homolog gene family, member B
|
|
chr22_+_48966394
|
0.358
|
|
TRABD
|
TraB domain containing
|
|
chr21_+_45649429
|
0.357
|
NM_130445
|
COL18A1
|
collagen, type XVIII, alpha 1
|
|
chr13_-_74953873
|
0.345
|
NM_014832
|
TBC1D4
|
TBC1 domain family, member 4
|
|
chr16_+_18986417
|
0.340
|
NM_016138
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr16_+_18986721
|
0.336
|
NM_001190983
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chrX_+_46825179
|
0.335
|
|
RGN
|
regucalcin (senescence marker protein-30)
|
|
chr17_+_77528699
|
0.329
|
NM_024083
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1
|
|
chr5_-_32210084
|
0.316
|
|
GOLPH3
|
golgi phosphoprotein 3 (coat-protein)
|
|
chr17_-_20311411
|
0.313
|
NM_001042685
|
LGALS9B
|
lectin, galactoside-binding, soluble, 9B
|
|
chr3_-_49117169
|
0.305
|
NM_005051
|
QARS
|
glutaminyl-tRNA synthetase
|
|
chr18_-_6404900
|
0.304
|
NM_173464
|
L3MBTL4
|
l(3)mbt-like 4 (Drosophila)
|
|
chr17_-_59451662
|
0.288
|
NM_001099786
NM_001099787
NM_001099788
NM_001099789
|
ICAM2
|
intercellular adhesion molecule 2
|
|
chr22_+_48966480
|
0.286
|
NM_025204
|
TRABD
|
TraB domain containing
|
|
chr17_+_77528782
|
0.277
|
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1
|
|
chr19_-_1601246
|
0.274
|
NM_001136139
|
TCF3
|
transcription factor 3 (E2A immunoglobulin enhancer binding factors E12/E47)
|
|
chr10_+_80569464
|
0.271
|
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr21_-_45165249
|
0.270
|
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit)
|
|
chr5_-_32210154
|
0.270
|
NM_022130
|
GOLPH3
|
golgi phosphoprotein 3 (coat-protein)
|
|
chr5_-_32210142
|
0.265
|
|
GOLPH3
|
golgi phosphoprotein 3 (coat-protein)
|
|
chr22_+_16973556
|
0.263
|
|
TUBA8
|
tubulin, alpha 8
|
|
chr5_-_32210112
|
0.262
|
|
GOLPH3
|
golgi phosphoprotein 3 (coat-protein)
|
|
chr14_+_100262943
|
0.253
|
NM_003836
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr19_+_50100843
|
0.250
|
NM_000041
|
APOE
|
apolipoprotein E
|
|
chr19_-_9592883
|
0.249
|
NM_152289
|
ZNF561
|
zinc finger protein 561
|
|
chr11_+_36572615
|
0.246
|
NM_138787
|
C11orf74
|
chromosome 11 open reading frame 74
|
|
chr5_-_32209999
|
0.238
|
|
GOLPH3
|
golgi phosphoprotein 3 (coat-protein)
|
|
chr7_+_128365226
|
0.237
|
NM_001098629
NM_001098630
|
IRF5
|
interferon regulatory factor 5
|
|
chr22_+_16973399
|
0.235
|
NM_001193414
NM_018943
|
TUBA8
|
tubulin, alpha 8
|
|
chr14_-_34943574
|
0.230
|
NM_020529
|
NFKBIA
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha
|
|
chr2_-_39517795
|
0.227
|
|
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3
|
|
chr1_+_181259317
|
0.227
|
|
|
|
|
chr10_+_30763056
|
0.226
|
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8
|
|
chr8_-_93184629
|
0.220
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr11_+_7491789
|
0.218
|
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2)
|
|
chr21_-_45165204
|
0.211
|
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit)
|
|
chr19_-_48700824
|
0.210
|
NM_198850
|
PHLDB3
|
pleckstrin homology-like domain, family B, member 3
|
|
chr2_+_230495439
|
0.210
|
NM_174899
|
FBXO36
|
F-box protein 36
|
|
chr21_+_45649719
|
0.209
|
|
|
|
|
chr22_+_21464980
|
0.205
|
|
|
|
|
chr6_-_7855125
|
0.204
|
NM_001145549
|
TXNDC5
|
thioredoxin domain containing 5 (endoplasmic reticulum)
|
|
chr2_+_108271526
|
0.200
|
NM_001056
NM_176825
|
SULT1C2
|
sulfotransferase family, cytosolic, 1C, member 2
|
|
chr19_+_10652025
|
0.198
|
|
|
|
|
chr2_+_46623396
|
0.196
|
|
RHOQ
|
ras homolog gene family, member Q
|
|
chr10_-_101831579
|
0.193
|
NM_001308
|
CPN1
|
carboxypeptidase N, polypeptide 1
|
|
chr10_+_111757867
|
0.191
|
|
ADD3
|
adducin 3 (gamma)
|
|
chr14_-_34661387
|
0.190
|
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma
|
|
chr1_+_110329224
|
0.189
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr16_+_55950195
|
0.186
|
NM_002990
|
CCL22
|
chemokine (C-C motif) ligand 22
|
|
chr19_-_54267932
|
0.183
|
NM_031886
|
KCNA7
|
potassium voltage-gated channel, shaker-related subfamily, member 7
|
|
chr17_-_74289901
|
0.182
|
NM_004762
NM_017456
|
CYTH1
|
cytohesin 1
|
|
chr14_-_33489708
|
0.181
|
NM_022073
|
EGLN3
|
egl nine homolog 3 (C. elegans)
|
|
chr1_+_202031373
|
0.178
|
NM_014827
|
ZC3H11A
|
zinc finger CCCH-type containing 11A
|
|
chr20_+_337346
|
0.178
|
|
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1
|
|
chr7_-_94863558
|
0.178
|
NM_000940
|
PON3
|
paraoxonase 3
|
|
chr1_+_202031425
|
0.177
|
|
ZC3H11A
|
zinc finger CCCH-type containing 11A
|
|
chr16_+_14634324
|
0.173
|
|
BFAR
|
bifunctional apoptosis regulator
|
|
chr11_+_66034652
|
0.166
|
NM_024649
|
BBS1
|
Bardet-Biedl syndrome 1
|
|
chr22_+_21543671
|
0.164
|
|
IGLV4-3
|
immunoglobulin lambda variable 4-3
|
|
chr20_+_46971618
|
0.163
|
NM_006420
|
ARFGEF2
|
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited)
|
|
chr6_+_31538934
|
0.161
|
NM_006674
|
HCP5
|
HLA complex P5
|
|
chr17_-_8006979
|
0.161
|
NM_014232
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2)
|
|
chr1_-_181826292
|
0.160
|
NM_000433
NM_001190789
NM_001190794
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr7_+_128365482
|
0.158
|
NM_032643
|
IRF5
|
interferon regulatory factor 5
|
|
chr14_+_57834962
|
0.152
|
NM_002892
NM_023000
NM_023001
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like)
|
|
chr4_-_2727548
|
0.148
|
NM_001161527
|
TNIP2
|
TNFAIP3 interacting protein 2
|
|
chr21_+_39099624
|
0.145
|
|
ETS2
|
v-ets erythroblastosis virus E26 oncogene homolog 2 (avian)
|
|
chr2_-_203444468
|
0.144
|
NM_138468
|
ICA1L
|
islet cell autoantigen 1,69kDa-like
|
|
chr12_+_68459015
|
0.144
|
NM_001024647
|
RAB3IP
|
RAB3A interacting protein (rabin3)
|
|
chr15_+_72861767
|
0.140
|
|
CSK
|
c-src tyrosine kinase
|
|
chr14_-_93926679
|
0.140
|
NM_001002235
NM_001002236
NM_001127700
NM_001127701
NM_001127702
NM_001127703
NM_001127704
NM_001127705
NM_001127706
NM_001127707
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr9_-_5823072
|
0.140
|
NM_024896
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1
|
|
chr1_+_202031445
|
0.139
|
|
ZC3H11A
|
zinc finger CCCH-type containing 11A
|
|
chr18_-_31178415
|
0.136
|
NM_006965
|
ZNF24
|
zinc finger protein 24
|
|
chr6_+_35335456
|
0.133
|
NM_003427
|
ZNF76
|
zinc finger protein 76
|
|
chr1_+_110328955
|
0.132
|
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr19_+_40223249
|
0.130
|
NM_002151
NM_182983
|
HPN
|
hepsin
|
|
chr16_+_14634168
|
0.129
|
NM_016561
|
BFAR
|
bifunctional apoptosis regulator
|
|
chr19_+_54962075
|
0.126
|
|
AP2A1
|
adaptor-related protein complex 2, alpha 1 subunit
|
|
chr9_+_20648307
|
0.125
|
NM_017794
|
KIAA1797
|
KIAA1797
|
|
chr19_-_4510713
|
0.124
|
|
SEMA6B
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B
|
|
chr20_+_3749164
|
0.123
|
NM_018347
|
C20orf29
|
chromosome 20 open reading frame 29
|
|
chr16_-_88295595
|
0.123
|
NM_152339
|
SPATA2L
|
spermatogenesis associated 2-like
|
|
chr12_-_6450015
|
0.120
|
NM_014231
NM_016830
NM_199245
|
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1)
|
|
chr19_-_47451119
|
0.119
|
NM_006494
|
ERF
|
Ets2 repressor factor
|
|
chr12_+_55896844
|
0.119
|
NM_007224
|
NXPH4
|
neurexophilin 4
|
|
chr15_+_89229275
|
0.117
|
NM_001143783
NM_001143784
|
FES
|
feline sarcoma oncogene
|
|
chr1_-_45924770
|
0.117
|
NM_021639
|
GPBP1L1
|
GC-rich promoter binding protein 1-like 1
|
|
chr11_+_63754328
|
0.117
|
NM_005528
|
DNAJC4
|
DnaJ (Hsp40) homolog, subfamily C, member 4
|
|
chr10_+_35524835
|
0.117
|
NM_182721
NM_182722
NM_182723
NM_182724
NM_182725
|
CREM
|
cAMP responsive element modulator
|
|
chr1_+_227473458
|
0.115
|
NM_004578
|
RAB4A
|
RAB4A, member RAS oncogene family
|
|
chr11_+_59979857
|
0.115
|
NM_021950
NM_152866
|
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1
|
|
chr11_+_17254854
|
0.115
|
NM_005013
|
NUCB2
|
nucleobindin 2
|
|
chrX_-_130251068
|
0.111
|
NM_001170961
NM_001170962
NM_001555
NM_205833
|
IGSF1
|
immunoglobulin superfamily, member 1
|
|
chr9_-_5822840
|
0.111
|
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1
|
|
chr17_+_69974375
|
0.108
|
|
CD300A
|
CD300a molecule
|
|
chr3_+_12367993
|
0.108
|
NM_015869
|
PPARG
|
peroxisome proliferator-activated receptor gamma
|
|
chr21_+_39099717
|
0.107
|
NM_005239
|
ETS2
|
v-ets erythroblastosis virus E26 oncogene homolog 2 (avian)
|
|
chr14_-_34661156
|
0.106
|
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma
|
|
chr4_+_1872150
|
0.106
|
NM_133334
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1
|
|
chr14_-_34660902
|
0.103
|
|
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma
|
|
chr15_-_41000100
|
0.103
|
NM_173500
|
TTBK2
|
tau tubulin kinase 2
|
|
chr7_-_38637475
|
0.102
|
|
AMPH
|
amphiphysin
|
|
chr1_+_149309703
|
0.102
|
NM_144618
|
GABPB2
|
GA binding protein transcription factor, beta subunit 2
|
|
chr19_-_47450991
|
0.101
|
|
ERF
|
Ets2 repressor factor
|
|
chr3_-_150952944
|
0.101
|
NM_016094
|
COMMD2
|
COMM domain containing 2
|
|
chr16_+_15950707
|
0.101
|
NM_004996
NM_019862
NM_019898
NM_019899
NM_019900
|
ABCC1
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 1
|
|
chr6_-_8009560
|
0.101
|
|
MUTED-TXNDC5
MUTED
|
MUTED-TXNDC5 read-through transcript
muted homolog (mouse)
|
|
chr3_+_52504410
|
0.100
|
|
STAB1
|
stabilin 1
|
|
chr22_+_45071301
|
0.099
|
NM_016426
|
GTSE1
|
G-2 and S-phase expressed 1
|
|
chr7_-_120285364
|
0.098
|
NM_012338
|
TSPAN12
|
tetraspanin 12
|
|
chrX_-_39921525
|
0.096
|
NM_001123383
NM_001123384
|
BCOR
|
BCL6 corepressor
|
|
chr12_-_6668085
|
0.094
|
NM_001039916
|
ZNF384
|
zinc finger protein 384
|
|
chr3_-_197643693
|
0.093
|
|
UBXN7
|
UBX domain protein 7
|
|
chr14_+_57835028
|
0.092
|
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like)
|
|
chr4_-_2727816
|
0.092
|
NM_024309
|
TNIP2
|
TNFAIP3 interacting protein 2
|
|
chr18_-_31178379
|
0.089
|
|
ZNF24
|
zinc finger protein 24
|
|
chr11_+_124115051
|
0.088
|
|
NRGN
|
neurogranin (protein kinase C substrate, RC3)
|
|
chr22_+_44084430
|
0.084
|
NM_017911
|
FAM118A
|
family with sequence similarity 118, member A
|
|
chr16_-_31013626
|
0.083
|
NM_024006
NM_206824
|
VKORC1
|
vitamin K epoxide reductase complex, subunit 1
|
|
chr3_-_54937001
|
0.083
|
NM_020678
|
LRTM1
|
leucine-rich repeats and transmembrane domains 1
|
|
chr3_-_197643669
|
0.082
|
|
UBXN7
|
UBX domain protein 7
|
|
chr11_-_65841077
|
0.082
|
NM_020404
|
CD248
|
CD248 molecule, endosialin
|
|
chr19_-_12305435
|
0.081
|
NM_145276
|
ZNF563
|
zinc finger protein 563
|
|
chr19_+_60878381
|
0.080
|
|
EPN1
|
epsin 1
|
|
chr6_-_79844606
|
0.078
|
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr16_-_3707450
|
0.078
|
NM_016292
|
TRAP1
|
TNF receptor-associated protein 1
|
|
chr1_-_44912704
|
0.077
|
|
TMEM53
|
transmembrane protein 53
|
|
chr3_+_153469421
|
0.077
|
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr15_+_67239853
|
0.076
|
NM_015554
|
GLCE
|
glucuronic acid epimerase
|
|
chr4_+_2764510
|
0.076
|
NM_001122681
|
SH3BP2
|
SH3-domain binding protein 2
|
|
chr3_-_197643701
|
0.073
|
NM_015562
|
UBXN7
|
UBX domain protein 7
|
|
chr16_-_31013550
|
0.073
|
|
VKORC1
|
vitamin K epoxide reductase complex, subunit 1
|
|
chr1_-_39929654
|
0.071
|
NM_016257
|
HPCAL4
|
hippocalcin like 4
|
|
chr12_+_112143658
|
0.071
|
|
TPCN1
|
two pore segment channel 1
|
|
chr17_+_7063873
|
0.071
|
NM_000018
NM_001033859
|
ACADVL
|
acyl-CoA dehydrogenase, very long chain
|
|
chr16_+_11978002
|
0.071
|
NM_032167
|
RUNDC2A
|
RUN domain containing 2A
|
|
chr15_+_57691272
|
0.070
|
NM_004751
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type
|
|
chr12_+_112143670
|
0.070
|
|
TPCN1
|
two pore segment channel 1
|
|
chr20_+_37024383
|
0.069
|
NM_001190809
NM_021931
|
DHX35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35
|
|
chr1_+_224803123
|
0.069
|
NM_001003665
|
C1orf95
|
chromosome 1 open reading frame 95
|
|
chr9_+_115396020
|
0.069
|
NM_144489
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr16_-_31013594
|
0.069
|
|
VKORC1
|
vitamin K epoxide reductase complex, subunit 1
|
|
chr7_+_39956633
|
0.069
|
|
CDK13
|
cyclin-dependent kinase 13
|
|
chr5_-_179166230
|
0.069
|
|
MGAT4B
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B
|
|
chr11_-_73700257
|
0.068
|
NM_182904
|
P4HA3
|
prolyl 4-hydroxylase, alpha polypeptide III
|
|
chr2_-_118488189
|
0.067
|
NM_019044
|
CCDC93
|
coiled-coil domain containing 93
|
|
chr16_+_1974182
|
0.066
|
|
GFER
|
growth factor, augmenter of liver regeneration
|
|
chr3_+_52504391
|
0.066
|
NM_015136
|
STAB1
|
stabilin 1
|
|
chr7_+_39956480
|
0.066
|
NM_003718
NM_031267
|
CDK13
|
cyclin-dependent kinase 13
|
|
chr4_-_122521562
|
0.064
|
NM_198179
|
QRFPR
|
pyroglutamylated RFamide peptide receptor
|
|
chr19_-_5854718
|
0.063
|
|
NDUFA11
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa
|
|
chr8_-_53789535
|
0.063
|
|
RB1CC1
|
RB1-inducible coiled-coil 1
|
|
chr9_+_35663852
|
0.063
|
NM_001216
|
CA9
|
carbonic anhydrase IX
|
|
chr6_+_31910623
|
0.063
|
NM_001040437
NM_001040438
|
C6orf48
|
chromosome 6 open reading frame 48
|
|
chr16_+_3447957
|
0.062
|
NM_001083600
NM_024845
|
NAT15
|
N-acetyltransferase 15 (GCN5-related, putative)
|
|
chr17_+_45008473
|
0.062
|
|
NXPH3
|
neurexophilin 3
|
|
chr19_+_11936868
|
0.061
|
NM_001012753
|
ZNF763
|
zinc finger protein 763
|
|
chr2_+_74595073
|
0.061
|
NM_016170
|
TLX2
|
T-cell leukemia homeobox 2
|
|
chr20_-_33463174
|
0.060
|
|
UQCC
|
ubiquinol-cytochrome c reductase complex chaperone
|
|
chr7_+_99451110
|
0.060
|
NM_003439
|
ZKSCAN1
|
zinc finger with KRAB and SCAN domains 1
|
|
chr15_-_73017439
|
0.057
|
NM_004255
|
COX5A
|
cytochrome c oxidase subunit Va
|
|
chr5_-_168204430
|
0.057
|
|
|
|
|
chr11_+_46595688
|
0.056
|
|
ATG13
|
ATG13 autophagy related 13 homolog (S. cerevisiae)
|
|
chr1_-_173979292
|
0.055
|
NM_003285
|
TNR
|
tenascin R (restrictin, janusin)
|
|
chr14_+_23654307
|
0.055
|
NM_181357
|
DCAF11
|
DDB1 and CUL4 associated factor 11
|
|
chr3_-_13032130
|
0.055
|
|
|
|
|
chr15_-_19974402
|
0.055
|
|
|
|
|
chr20_+_30192968
|
0.053
|
|
TM9SF4
|
transmembrane 9 superfamily protein member 4
|
|
chr16_+_27320983
|
0.053
|
NM_181078
NM_181079
|
IL21R
|
interleukin 21 receptor
|
|
chr19_+_44591589
|
0.053
|
|
|
|
|
chr1_-_44912649
|
0.052
|
NM_024587
|
TMEM53
|
transmembrane protein 53
|
|
chr16_+_31027231
|
0.052
|
|
BCKDK
|
branched chain ketoacid dehydrogenase kinase
|
|
chr5_-_63293301
|
0.052
|
NM_000524
|
HTR1A
|
5-hydroxytryptamine (serotonin) receptor 1A
|
|
chr20_+_39199263
|
0.051
|
|
PLCG1
|
phospholipase C, gamma 1
|
|
chr19_+_60878238
|
0.049
|
NM_001130072
NM_013333
|
EPN1
|
epsin 1
|
|
chr12_+_112143599
|
0.049
|
NM_001143819
NM_017901
|
TPCN1
|
two pore segment channel 1
|
|
chr19_-_52679287
|
0.048
|
NM_007059
|
KPTN
|
kaptin (actin binding protein)
|